Research Interests
Explore my research interests and innovative work products.
Scaffolding within the Nucleus
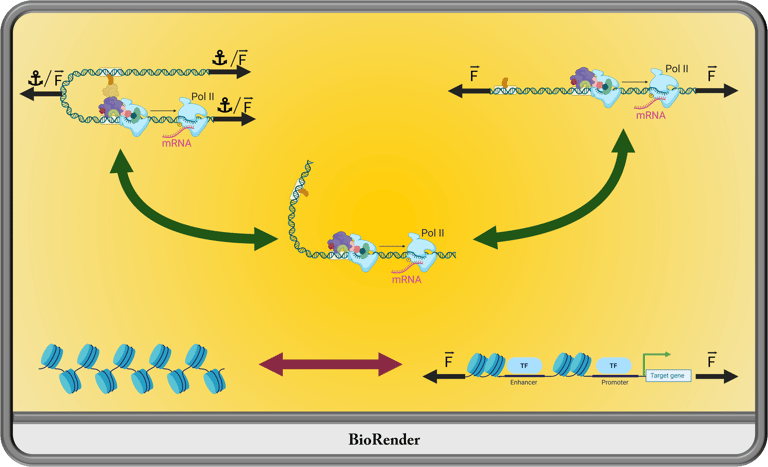
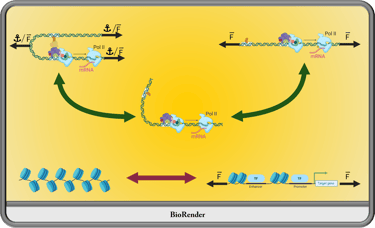
How do mechanical stimuli generated within and outside the nucleus contribute to genome architecture and gene regulation? We know mechanical forces are felt within the nucleus and we know there are mechanisms to remedy excesses in these mechanical transmissions, but little is known about how cell biology takes advantage of these cues to respond and organize. I plan to employ a combination of transgenic nanotools, advanced imaging techniques, chromatin capture technology, and unique model organism strategies to elucidate the epigenetic influences of mechanical stimuli in vivo.
Transgenic Tool Development
An ever growing number of biological questions have arisen which cannot be answered using traditional methods. To answer the call for tools to peak into the once-black box that is the currently functioning animal cell, a vast variety of genetically encoded nano-devices have been developed. Every tool of this kind has the potential to enrich entire fields of biological research by enabling the community to answer their own challenging questions independently. I intend to develop several transgenic tools designed to elucidate real time, subcellular mysteries within living organisms.
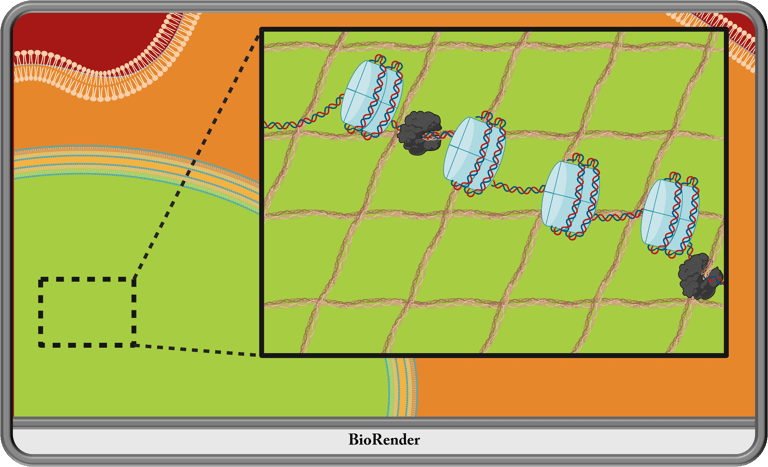
Since the nuclear f-actin wars at the turn of the century, investigating the existence and composition of the nucleoskeleton has been mired in controversy. However, recent inquiries, predominantly in the context of cancer and brain development research, have revealed a structural role of Matrin3 important for proper nuclear organization and gene expression. I plan to employ mouse transgenics, advanced imaging techniques, and sequencing techniques to lay this dispute to rest.
Mechanical Regulation of Chromatin Organizition
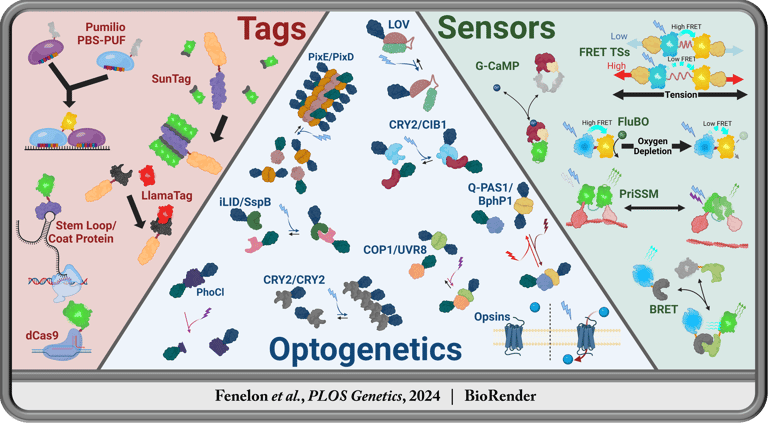
Past Research
Explore my research interests and innovative work products.
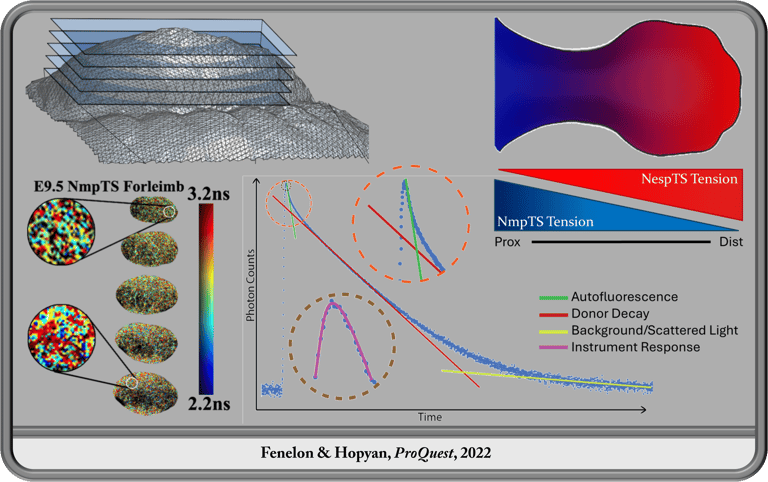
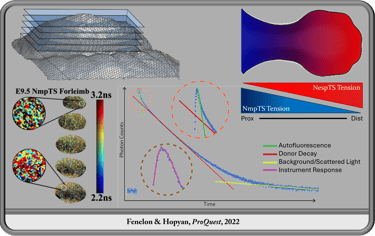
Transgenic Tools for Measuring Transnuclear Forces in Live Mice in vivo
The chromatin is the dominant structural component of the nucleus which is a heavily interconnected organelle linked through mechanical linkages through the cytoplasm and cell membrane to the extracellular membrane and cell-cell junctions providing clear epigenetic potential. During my PhD research in the Hopyan Lab, I was able to demonstrate the existence of an axial transcuclear force gradient onto the chromatin during forelimb bud morphogenesis. This novel discovery bares intriguing implications for mechanotransduction as an epigenetic mechanism during development whereby mechanical forces driven by morphogenetic shape changes and cell and tissue dynamics are transmitted to the nuclear interior to mechanically influence chromatin architecture and potentially gene regulation.
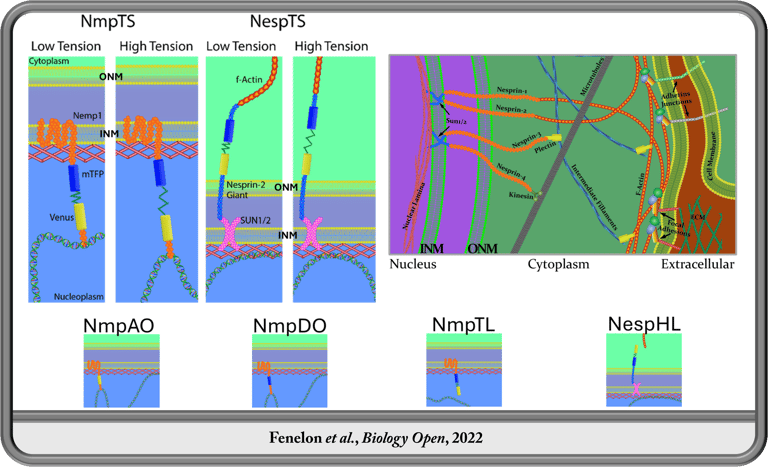
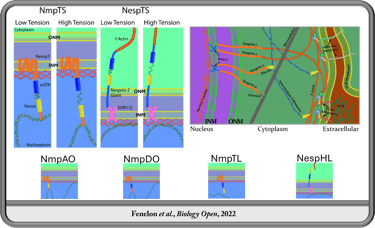
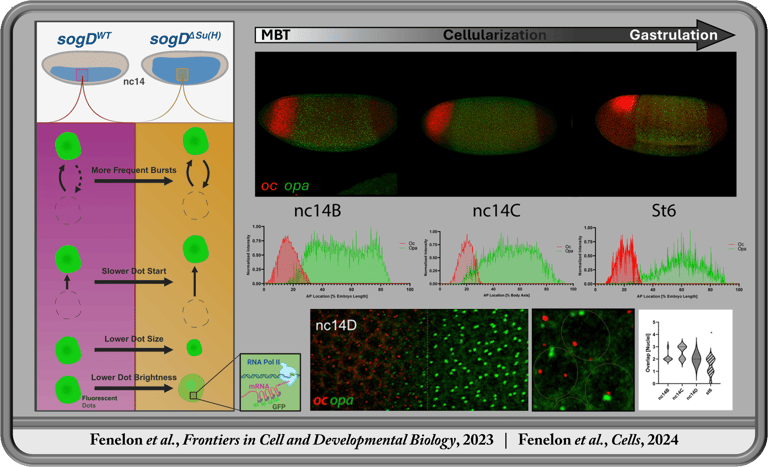
Spatiotemporal Transcription Factor Dynamics in Pre-Gastrulation Brain Formation
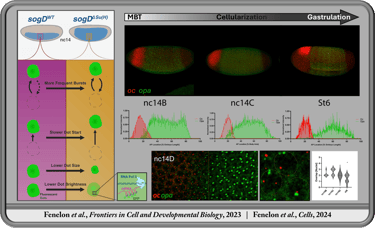
In the Koromila Lab, I investigated transcription factor spatiotemporal expression and regulatory dynamics during the transition from blastula to gastrula (stages 4-6). We demonstrated that opa and oc transiently co-occupy cells in a narrow band where the brain later forms prior to gastrulation onset. This region coincides with gene expression patterns of brain related genes and enhancers occupied by Opa and/or Oc. We further investigated the dynamic function of Su(H) during this transition, revealing an antinomious role between tissues whereby Su(H) facilitates activation of sog through the sog-Distal enhancer via modulation of transcriptional bursting ventrolaterally, contrary to its well established role in repression through the same regulatory element dorsolaterally prior to gastrulation.
In the Hopyan Lab, I designed and developed transgenic mouse models for measuring force transmission across the nuclear membrane through confocal live imaging. These tools, now available to the scientific community, are of particular significance to science as they function in mouse embryos which are a notoriously challenging model for microscopy endeavors. To meet this challenge, in addition to the transgenic animals themselves, we developed an analysis software for fitting FLIM data from sources with high background light and autofluorescence, FLIMvivo, which is now available to the scientific community and useful for analysis of similar data from other model organisms or in vitro cultures as well as challenging organisms like the mouse embryo.
Elucidating the Role of Nuclear Mechanotransduction During Embryonic Development
Current Research
Check back here for updates as I embark on a new chapter in my journey to achieving my own lab.